daptics Validation
daptics gets results. Three examples
of successful application of daptics to experimental campaigns in the
laboratory are described below. Many other kinds of complex discovery
and optimization problems can be solved by daptics.
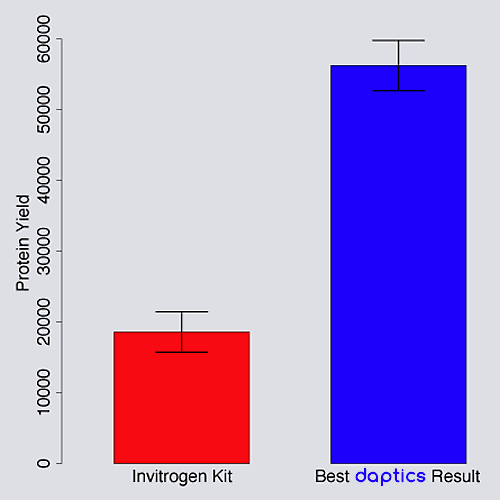
In-vitro protein synthesis
daptics has been used to optimize the amount of functional protein synthesized with a commercial in vitro synthesis kit.3 The experimental parameters included many possible DNA sequences, amino acid proportions, salts, buffers, and process variables.
This was a huge experimental space, and systematically exploring every experiment (over 1.5 million) would have taken an enormous amount of time and would have been extremely expensive.
daptics increased protein yield by 350% after exploring
less than 0.015% of the experimental space.4
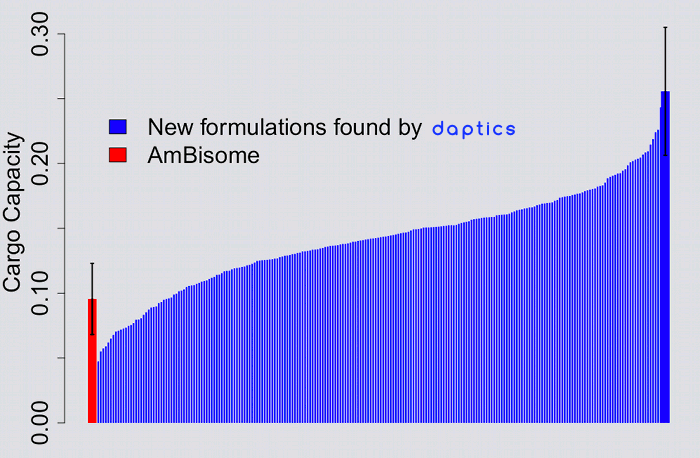
Drug formulations
AmBisome™ is the leading formulation of Amphotericin B, an anti-fungal drug. daptics was applied to find novel formulations of the drug, optimized for cargo capacity.
A variety of amphiphilic molecules, salts, buffers, and other components made up an experimental space of over 80,000 experiments.
By exploring less than 0.6% of the possible experiments, daptics found
hundreds of novel formulations, many of which were very stable
and substantially increased cargo capacity.2
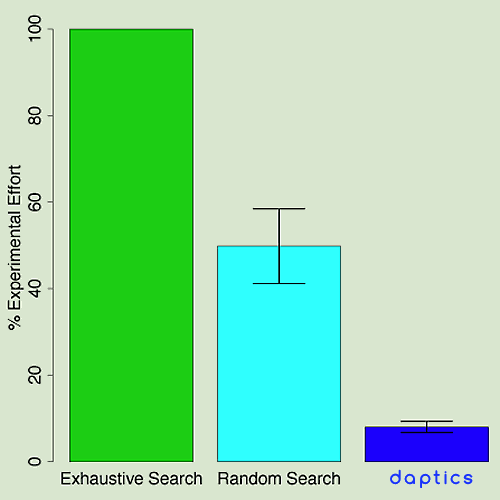
Synergistic drug combinations
A pharmaceutical company, CombinatoRx, exhaustively screened many
combinations of drugs, looking for synergistic therapeutic
effects.5 This took months of time and hundreds of
thousands of individual experiments. When applied to the data from
this experiment, daptics found the optimal combination in the experimental
space with less than 10% of the experimental effort.
daptics's Power
Simulations are a powerful and cost-effective way to demonstrate how
efficiently daptics finds optimal targets
in complex search spaces. The following tests were run on simulated
but realistic experimental search spaces, representing combinations of
a certain number of experimental parameters (e.g., materials or compounds)
chosen from a library of 100, with each parameter in the combination
taking any of 20 different values.1 The response
surfaces for these systems contained several local optima with a range of values
measured in arbitrary units from 0 to 8, with added experimental
noise. These simulations benchmark daptics against established
optimization methods in large and highly synergistic experimental
search spaces, and demonstrate daptics's superiority at identifying the
optimal targets while saving time and resources.
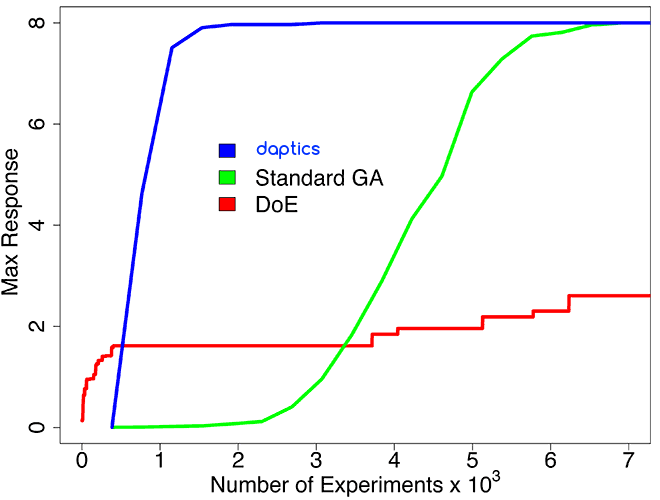
Pairwise combinations
A simulated system with pairwise combinations was defined on an experimental space with a total of 105 experiments. Exhaustive exploration of such a space in the laboratory would require a vast effort, and screening even a fraction of it could be very expensive (e.g., if the experimental parameters were costly pharmaceuticals or other chemicals).
On this system, standard Design of Experiments (DoE) techniques were obviously more efficient than exhaustive exploration, but substantially less efficient than daptics.
daptics found all of the optimal targets after exploring only 3% of the
experimental space, and was more than twice as efficient in doing so than a
standard genetic algorithm (GA).
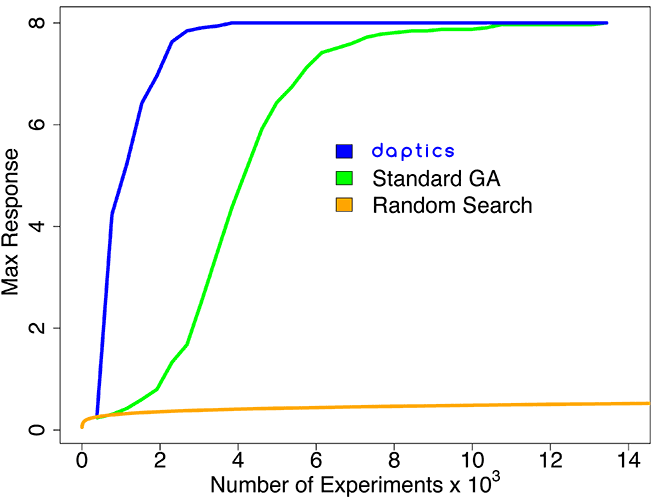
Higher-order (five-way) combinations
This simulated system involved all combinations of up to five experimental parameters, for a huge experimental space of over 1011 experiments. This system is more realistic than the one described above, because unpredictable higher-order synergies often occur in real-world systems: e.g., chemical systems for protein crystallization, protein synthesis, combination drug discovery, formulation of small molecule drugs, complex formulations (such as siRNA), heterogeneous catalyst discovery, and plastic formulations.
Because of such synergies, standard DoE cannot be used effectively to find optimal targets; DoE software such as JMP™ performs only slightly better than random search in a complex space of these dimensions.
daptics found all of the optimal targets in this space after exploring only 4,000 experiments, and was over 3 times more efficient than a standard GA.
1 Cawse J., Gazzola G., and Packard, N. (2011). Efficient discovery and optimization of complex high-throughput experiments. Catalysis today, 159(1): 55-63.
2 Caschera F., Gazzola G., Bedau M. A., Moreno C., Buchanan A., Cawse J., Packard N. H., and Hanczyc M. M. (2010). Automated discovery of novel drug formulations using predictive iterated high-throughput experimentation. PLoS One, 5(1):e8546.
3 Invitrogen Expressway; Cell-Free E. coli Expression System.
4 Caschera F., Bedau M. A., Buchanan A., Cawse J., de Lucrezia D., Gazzola G., Hanczyc M. M., and Packard N. H. (2011). Coping with complexity: Machine learning optimization of cell-free protein synthesis. Biotechnology and Bioengineering, 108(9): 2218-28.
5 Lehar, J. et al. (2007). Chemical combination effects predict connectivity in biological systems. Molecular Systems Biology, 3(80): 1-14.